Is the 5,10methylenetetrahydrofolate cofactor synthesized through a non-enzymatic or enzymatic mechanism?
Fernandes, H. S., Sousa, S. F., and Cerqueira, N. M. F. S. A.

My personal webpage
Fernandes, H. S., Sousa, S. F., and Cerqueira, N. M. F. S. A.

On September 28-29, I was presenting my recently published work at the UCIBIO Annual Meeting in Lisbon (FCT-NOVA).
Henrique S. Fernandes, M. J. Ramos, Sérgio F. Sousa, and N. M. F. S. A. Cerqueira
Centro Cultural Vila Flor, Guimarães
2016, 22th to 24th April
Poster: “Computational studies addressed to the catalytic mechanism of Histidine Decarboxylase”
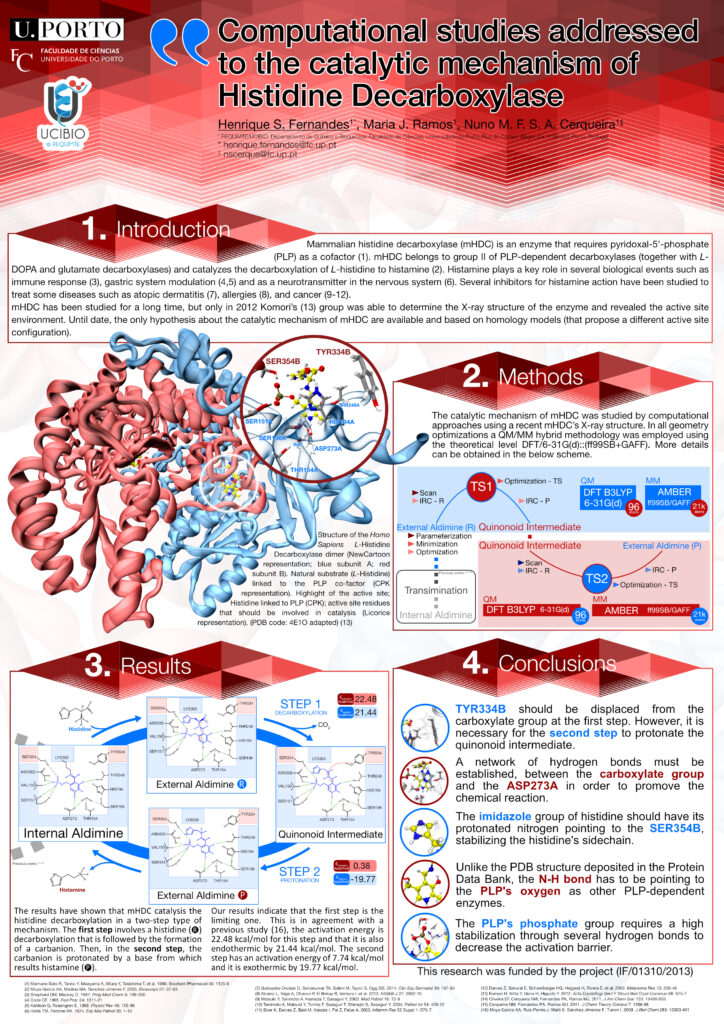
Henrique S. Fernandes (1), Maria João Ramos, Nuno M. F. S. A. Cerqueira (1)
(1) UCIBIO/REQUIMTE, Departamento de Química e Bioquímica, Faculdade de Ciências, Universidade do Porto, Rua do Campo Alegre s/n, 4169-007 Porto, PT
Mammalian histidine decarboxylase (mHDC) is an enzyme that requires pyridoxal-5′-phosphate (PLP) as a cofactor [1-3]. mHDC belongs to the group II of PLP-dependent decarboxylases together with L-DOPA and glutamate decarboxylases, and catalyses the L-histidine decarboxylation from which results histamine.
Histamine plays a key role in several biological events such as immune response, gastric system modulation and as a neurotransmitter in the nervous system. Several inhibitors for histamine action have been studied in order to treat some diseases such as atopic dermatitis, allergies, and cancer.
mHDC has been studied for a long time, but only in 2012 Komori’s [2] group was able to determine the X-ray structure of the enzyme and revealed the active site environment. Till date, only hypothesis about the catalytic mechanism of mHDC were available and based on homology models (that propose a different active site configuration).
In this work, we studied the catalytic mechanism of mHDC by computational approaches using the recent X-ray structure of mHDC (PDB code: 4E1O [4]) and a QM/MM methodology.
The results have shown that mHDC catalyses the reaction in a two-step type of mechanism. The first step involves a decarboxylation that is followed by the formation of a stable carbanion. In the second step, the carbanion is protonated by a base from which results histamine.
[1] Ngo, H. P., Cerqueira, N. M., Kim, J. K., Hong, M. K., Fernandes, P. A., Ramos, M. J., and Kang, L. W., Acta crystallographica. Section D, Biological crystallography 2014, 70, 596-606
[2] Oliveira, Eduardo F.; Cerqueira, Nuno M. F. S. A.; Fernandes, Pedro A. and Ramos, M.J., Journal of the American Chemical Society 2011, 133, 15496-15505
[3] Cerqueira, N. M. F. S. A.; Fernandes, P. A.; Ramos, M. J., Journal of Chemical Theory and Computation 2011, 7, 1356-1368
[4] Komori, H., Nitta, Y., Ueno, H., and Higuchi, Y., Acta Crystallogr Sect F Struct Biol Cryst Commun 2012, 68, 675-677
Instituto Pedro Nunes, Coimbra
2015, 18th December
Poster: “Computational studies addressed to the catalytic mechanism of Histidine Decarboxylase”
Henrique S. Fernandes (1), Nuno M. F. S. A. Cerqueira (1)
(1) UCIBIO/REQUIMTE, Departamento de Química e Bioquímica, Faculdade de Ciências, Universidade do Porto, Rua do Campo Alegre s/n, 4169-007 Porto, PT
Mammalian histidine decarboxylase (mHDC) is an enzyme that requires pyridoxal-5′-phosphate (PLP) as a cofactor [1]. mHDC belongs to the group II of PLP-dependent decarboxylases together with L-DOPA and glutamate decarboxylases, and catalyses the L-histidine decarboxylation from which results histamine.
Histamine plays a key role in several biological events such as immune response, gastric system modulation and as a neurotransmitter in the nervous system. Several inhibitors for histamine action have been studied in order to treat some diseases such as atopic dermatitis, allergies, and cancer.
mHDC has been studied for a long time, but only in 2012 Komori’s [2] group was able to determine X-ray structure of the enzyme and revealed the active site environment. Until date, only hypothesis about the mechanism of mHDC were available and based on homology models (that propose a different active site configuration).
In this work we are studying the catalytic mechanism of mHDC by computational means using the recent X-ray structure of mHDC and a QM/MM methodology.
The results have shown that mHDC catalyses the reaction in a two-step type of mechanism. The first step involves a decarboxylation that is followed by the formation of a carbanion. In the second step, the carbanion is protonated by a base from which results histamine. Our early results indicate that the first step is the limiting reaction step and the full reaction is endothermic by approximately 25 kcal/mol.
[1] Ngo HP, Cerqueira NMFSA, Kim JK, Hong MK, Fernandes PA, et al. 2014. Acta Crystallogr D Biol Crystallogr 70: 596-606;
[2] Komori H, Nitta Y, Ueno H, Higuchi Y. 2012. Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 675-7