Utilização de software na simulação molecular
Inf F0-I1
Nuno Cerqueira, Sérgio Sousa, Henrique Fernandes e Carla Teixeira- Investigadores do DQB-FCUP
My personal webpage
Inf F0-I1
Nuno Cerqueira, Sérgio Sousa, Henrique Fernandes e Carla Teixeira- Investigadores do DQB-FCUP
Dia 9 de Julho de 2018, 14h30
Auditório Grande do Centro Cultural Vila Flor
Maria João Ramos, Pedro Alexandrino Fernandes e Henrique Fernandes – Departamento de Química e Bioquímica da Faculdade de Ciências da Universidade do Porto
The molUP plugin, developed by Nuno Sousa Cerqueira and colleagues and described on page 1344, overcomes some of the most common problems in computational chemistry concerning the analysis of big data. MolUP was developed for use in the analysis of quantum chemistry (QM), QM/MM (molecular mechanics), and QM/QM calculations, molecular dynamics (MD) simulations, as well as the preparation of input files. MolUP also provides new tools to analyze and visualize existing computational chemistry information in a more userfriendly way that simplifies the current complex and time‐demanding practices used in the field. (DOI: 10.1002/jcc.25189)
CHEMTECH DQB – Chemistry Technology Events at DQB – 2018
March 8th, 2018 – Departamento de Química e Bioquímica da Faculdade de Ciências da Universidade do Porto

Realizou-se no Departamento de Química e Bioquímica da Faculdade de Ciências da Universidade do Porto uma mini feira de ciência onde se mostraram os softwares desenvolvidos no grupo de Bioquímica Teórica e Computacional. Os softwares são todos eles extensões para o VMD (Visual Molecular Dynamics) um visualizador molecular gratuito desenvolvido pela Universidade Illinois.
Os participantes tiveram a oportunidade de falar com os desenvolvedores e experimentar os softwares, que sendo gratuitos podem ser mais tarde instalados e usados por cada um dos participantes.
Fica aqui o folheto que foi distribuído na sessão demonstrativa: Download
E ainda algumas fotografias do evento:
André F. Pina
Carla S. S. Teixeira
Henrique S. Fernandes
Juliana F. Rocha
Nuno M. F. S. A. Cerqueira
Sérgio F. Sousa
Henrique Silva Fernandes, Maria João Ramos, and Nuno M.F.S.A. Cerqueira
Published on 21st February 2018
DOI: http://dx.doi.org/10.1002/jcc.25189 | Download citation
The notable advances obtained by computational (bio)chemistry provided its widespread use in many areas of science, in particular, in the study of reaction mechanisms. These studies involve a huge number of complex calculations, which are often carried out using the Gaussian suite of programs. The preparation of input files and the analysis of the output files are not easy tasks and often involve laborious and complex steps. Taking this into account, we developed molUP: a VMD plugin that offers a complete set of tools that enhance the preparation of QM and ONIOM (QM/MM, QM/QM, and QM/QM/MM) calculations. The starting structures for these calculations can be imported from different chemical formats. A set of tools is available to help the user to examine or modify any geometry parameter. This includes the definition of layers in ONIOM calculations, choosing fixed atoms during geometry optimizations, the recalculation or adjustment of the atomic charges, performing SCANs or IRC calculations, etc. molUP also extracts the geometries from the output files as well as the energies of each of them. All of these tasks are performed in an interactive GUI that is extremely helpful for the user. MolUP was developed to be easy to handle by inexperienced users, but simultaneously to be a fast and flexible graphical interface to allow the advanced users to take full advantage of this plugin. The program is available, free of charges, for macOS, Linux, and Windows at the PortoBioComp page https://www.fc.up.pt/PortoBioComp/database/doku.php?id=molup.
No dia 13 de agosto de 2017, o grupo de Bioquímica Teórica e Computacional esteve representado na Feira da 79ª Volta a Portugal em Bicicleta, no âmbito do projeto Volta ao Conhecimento promovido pelo Ministério da Ciência, Tecnologia e Ensino Superior.
O stand esteve montado junto à meta da 8ª etapa em Oliveira de Azeméis e contou com a participação de vários cidadãos que se deslocaram para assistir ao término de mais uma etapa da Volta.

Optámos por montar umm stand muito interativo com uma máquina de jogos educativa, o ProteinWars, cujo o objetivo é mostrar de que forma estudamos as proteínas e as moléculas para desenvolver novos fármacos.
Além disso, um segundo computador com um joystick acoplado permitiu aos participantes visualizarem moléculas e interagirem com os átomos em simulações moleculares.
Ver entrevista (Minuto 24:53)

Nanostructures for Cancer Therapy discusses the available preclinical and clinical nanoparticle technology platforms and their impact on cancer therapy, including current trends and developments in the use of nanostructured materials in chemotherapy and chemotherapeutics.
In particular, coverage is given to the applications of gold nanoparticles and quantum dots in cancer therapies. In addition to the multifunctional nanomaterials involved in the treatment of cancer, other topics covered include nanocomposites that can target tumoral cells and the release of antitumoral therapeutic agents.
The book is an up-to-date overview that covers the inorganic and organic nanostructures involved in the diagnostics and treatment of cancer.
Carla Teixeira*, Henrique Fernandes*, P. A. Fernandes, M. J. Ramos and Nuno M. F. S. A. Cerqueira (* contributed equally to this work)
Buy on Elsevier | Buy on Amazon | Available on iTunes | Preview

A growing understanding of tumor biology has allowed the identification of various cellular characteristics that are more frequently associated with cancer cells than with normal cells. These findings have prompted the development of new therapeutics specifically designed to exploit these differences. In this context, the amino acid depriving enzymes have shown very promising results and proven to be active and very specific against various types of cancers. These therapies involve the depletion of specific amino acids in the bloodstream that cannot be synthesized by tumor cells. This happens because these cells often have a defecting enzymatic armamentarium and therefore rely on external supply for those amino acids. Decreasing the concentration of certain amino acids in blood has thus been shown to impair the development or even destroy tumor cells. Normal cells remain unaltered since they are less demanding and/or can synthesize these compounds in sufficient amounts by other mechanisms.
In this chapter, the structure, function, catalytic mechanism and therapeutic application of some amino acid depriving enzymes will be reviewed. Particular attention will be given to enzymes that have potential or are currently used in the treatment of several types of cancer, namely: (1) l-asparaginase used for the treatment of acute lymphoblastic leukemia; (2) l-arginase and l-arginine deiminase that are used in the therapy of hepatocellular carcinomas and melanomas, two diseases that account annually with approximately 1 million of new cases and for which there is currently no efficacious treatment; and (3) l-Methioninase with potential to be used in the treatment of breast, colon, lung, and renal cancers.
“Moléculas que comandam a nossa vida” foi o nome dado à exposição de ilustrações científicas que decorreu, entre os dias 14 e 24 de março de 2017, na Galeria dos Leões da Reitoria da Universidade do Porto.
[googlemaps https://www.google.com/maps/embed?pb=!1m0!3m2!1spt-PT!2spt!4v1490604280961!6m8!1m7!1sF%3A-XeXy1JRnz_4%2FWMbSFwMc34I%2FAAAAAAAAE0Y%2F8cGV9kCABkoNvWJbnP3f7JbOI5HDCYgugCLIB!2m2!1d41.1469683!2d-8.6160073!3f290.56522785294374!4f2.9291030223811845!5f0.7820865974627469&w=600&h=450]
Centro Cultural Vila Flor, Guimarães
2016, 22th to 24th April
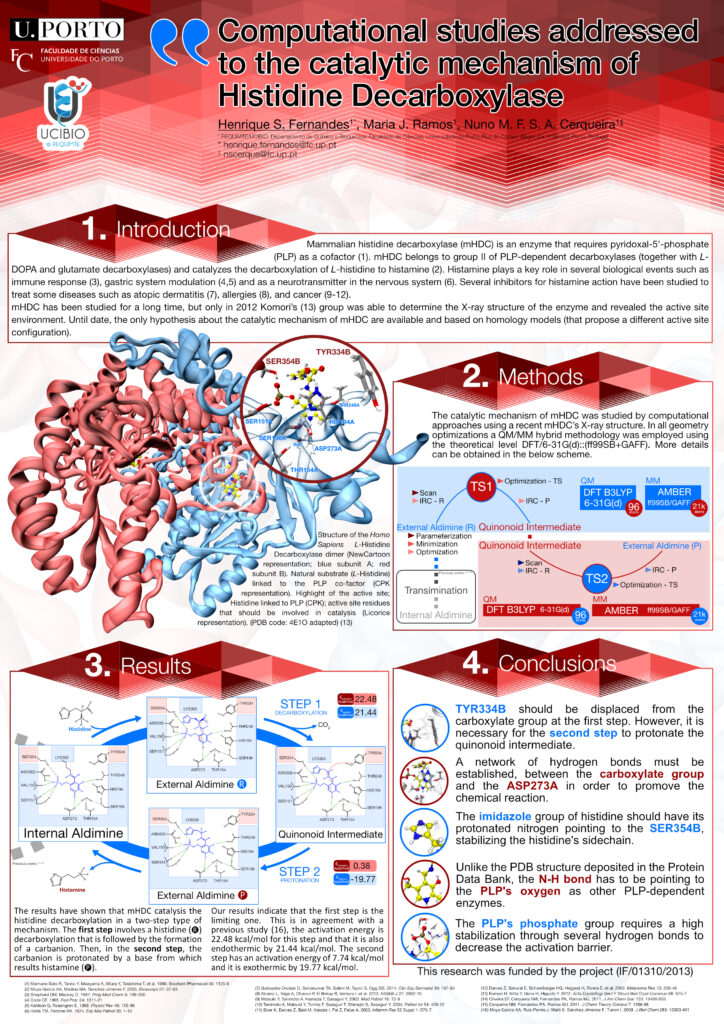
Poster: “Computational studies addressed to the catalytic mechanism of Histidine Decarboxylase”
Henrique S. Fernandes (1), Maria João Ramos, Nuno M. F. S. A. Cerqueira (1)
(1) UCIBIO/REQUIMTE, Departamento de Química e Bioquímica, Faculdade de Ciências, Universidade do Porto, Rua do Campo Alegre s/n, 4169-007 Porto, PT
Mammalian histidine decarboxylase (mHDC) is an enzyme that requires pyridoxal-5′-phosphate (PLP) as a cofactor [1-3]. mHDC belongs to the group II of PLP-dependent decarboxylases together with L-DOPA and glutamate decarboxylases, and catalyses the L-histidine decarboxylation from which results histamine.
Histamine plays a key role in several biological events such as immune response, gastric system modulation and as a neurotransmitter in the nervous system. Several inhibitors for histamine action have been studied in order to treat some diseases such as atopic dermatitis, allergies, and cancer.
mHDC has been studied for a long time, but only in 2012 Komori’s [2] group was able to determine the X-ray structure of the enzyme and revealed the active site environment. Till date, only hypothesis about the catalytic mechanism of mHDC were available and based on homology models (that propose a different active site configuration).
In this work, we studied the catalytic mechanism of mHDC by computational approaches using the recent X-ray structure of mHDC (PDB code: 4E1O [4]) and a QM/MM methodology.
The results have shown that mHDC catalyses the reaction in a two-step type of mechanism. The first step involves a decarboxylation that is followed by the formation of a stable carbanion. In the second step, the carbanion is protonated by a base from which results histamine.
[1] Ngo, H. P., Cerqueira, N. M., Kim, J. K., Hong, M. K., Fernandes, P. A., Ramos, M. J., and Kang, L. W., Acta crystallographica. Section D, Biological crystallography 2014, 70, 596-606
[2] Oliveira, Eduardo F.; Cerqueira, Nuno M. F. S. A.; Fernandes, Pedro A. and Ramos, M.J., Journal of the American Chemical Society 2011, 133, 15496-15505
[3] Cerqueira, N. M. F. S. A.; Fernandes, P. A.; Ramos, M. J., Journal of Chemical Theory and Computation 2011, 7, 1356-1368
[4] Komori, H., Nitta, Y., Ueno, H., and Higuchi, Y., Acta Crystallogr Sect F Struct Biol Cryst Commun 2012, 68, 675-677